Data Transformation
and Exploration
Rei Sanchez-Arias, Ph.D.
Using the dplyr package
Common tasks
Often you will need to create some new variables or summaries, or maybe you just want to rename the variables or reorder the observations to make the data a little easier to work with.
We will focus on how to use the
dplyrpackage, another core member of thetidyverse.
Prerequisites: Install the nycflights13 and tidyverse packages
library(tidyverse)library(nycflights13)Data from nycflights13
This dataset contains flights departing New York City (NYC) in 2013. It contains all 336,776 flights that departed from NYC in 2013.
The data comes from the US Bureau of Transportation Statistics, and is documented in ?flights
The tidyverse
The tidyverse is an opinionated collection of R packages designed for data science. All packages share an underlying design philosophy, grammar, and data structures.
The tidyverse
The tidyverse is an opinionated collection of R packages designed for data science. All packages share an underlying design philosophy, grammar, and data structures.
Definition of tidy data: (see paper by Hadley Wickham)
Each variable is a column
Each observation is a row
Each type of observational unit is a table
dplyr basics

Pick observations by their values:
filter()Reorder the rows:
arrange()Pick variables by their names:
select()Create new variables with functions of existing variables:
mutate()Collapse many values down to a single summary:
summarize()Operate on a group-by-group basis:
group_by()
Filtering rows
filter() allows you to subset observations based on their values. For example, we can select all flights on January 1st with:
filter(flights, month == 1, day == 1)Below is the dep_time and sched_dep_time for the first 100 rows
Comparisons
dplyr functions never modify their inputs, so if you want to save the result, you will need to use the assignment operator, <-
jan1 <- filter(flights, month == 1, day == 1)Comparisons
dplyr functions never modify their inputs, so if you want to save the result, you will need to use the assignment operator, <-
jan1 <- filter(flights, month == 1, day == 1)
To use filtering effectively, you have to know how to select the observations that you want using comparison operators:
>, >=, <, <=,
!= (not equal), and
== (equal).
Logical operators
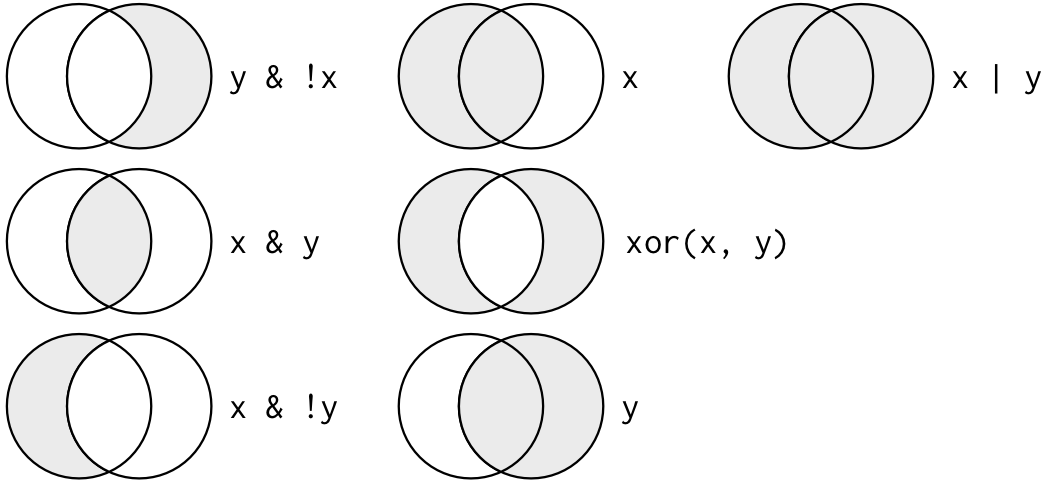
Flights in November OR December
The following code finds all flights that departed in November OR December:
filter(flights, month == 11 | month == 12)Flights in November OR December
The following code finds all flights that departed in November OR December:
filter(flights, month == 11 | month == 12)A useful short-hand is x %in% y. This will select every row where x is one of the values in y. We could use it to rewrite the code above:
nov_dec <- filter(flights, month %in% c(11, 12))Arrange rows with arrange()
arrange() works similarly to filter() except that instead of selecting rows, it changes their order.
It takes a data frame and a set of column names to order by.
arrange(flights, year, month, day)Arrange rows with arrange()
arrange() works similarly to filter() except that instead of selecting rows, it changes their order.
It takes a data frame and a set of column names to order by.
arrange(flights, year, month, day)Use desc() to re-order by a column in descending order:
arrange(flights, desc(arr_delay))Select columns with select()
select() allows you to rapidly zoom in on a useful subset using the names of the variables.
# Select columns by nameselect(flights, year, month, day)
There are a number of helper functions you can use within select():
starts_with("abc"): matches names that begin with "abc".
ends_with("xyz"): matches names that end with "xyz".
contains("ijk"): matches names that contain "ijk".
Add new variables with mutate()
To add new columns that are functions of existing columns. That is the job of mutate(). mutate() always adds new columns at the end of your dataset.
# create a smaller dataset with less columnsflights_sml <- select(flights, year:day, ends_with("delay"), distance, air_time)Example: using mutate()
mutate(flights_sml, gain = arr_delay - dep_delay, speed = distance / air_time * 60)Note that you can refer to columns that you have just created:
mutate(flights_sml, gain = arr_delay - dep_delay, hours = air_time / 60, gain_per_hour = gain / hours)A random sample of 100 observations is shown below:
| year | month | day | dep_delay | arr_delay | distance | air_time | gain | hours | gain_per_hour |
|---|
year | month | day | dep_delay | arr_delay | distance | air_time | gain | hours | gain_per_hour | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2013 | 9 | 9 | 10 | -9 | 199 | 37 | -19 | 0.616666666666667 | -30.8108108108108 |
| 2 | 2013 | 12 | 29 | -5 | -8 | 1096 | 174 | -3 | 2.9 | -1.03448275862069 |
| 3 | 2013 | 10 | 28 | -2 | -13 | 200 | 46 | -11 | 0.766666666666667 | -14.3478260869565 |
| 4 | 2013 | 5 | 7 | -3 | -22 | 762 | 101 | -19 | 1.68333333333333 | -11.2871287128713 |
| 5 | 2013 | 5 | 16 | -1 | -4 | 1605 | 220 | -3 | 3.66666666666667 | -0.818181818181818 |
| 6 | 2013 | 11 | 21 | 0 | -10 | 1028 | 138 | -10 | 2.3 | -4.34782608695652 |
Grouped summaries with summarise()
summarise() collapses a data frame to a single row:
summarise(flights, delay = mean(dep_delay, na.rm = TRUE))## # A tibble: 1 × 1## delay## <dbl>## 1 12.6The summarise() function is useful when we pair it with group_by().
This way, the analysis can be done for individual groups.
Example
The pipe sends the output of the LHS function to the first argument of the RHS function.
For example:
# pipe examplesum(1:8) %>% sqrt() %>% log()## [1] 1.791759is equivalent to
# the log of the square root# of the sum of the elements # of the vector [1 ... 8]log(sqrt(sum(1:8)))## [1] 1.791759Example for data exploration
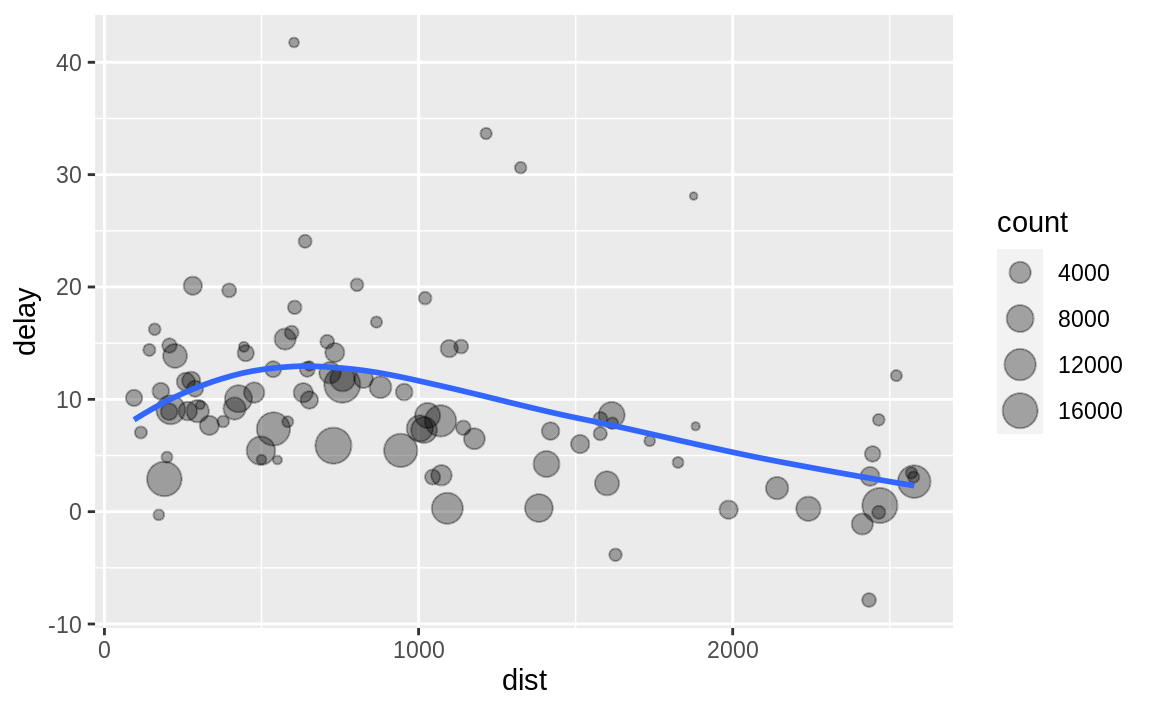
Imagine that we want to explore the relationship between the distance and average delay for each location.
Example for data exploration
Imagine that we want to explore the relationship between the distance and average delay for each location.
There are three steps to prepare this data:
Group flights by destination.
Summarize to compute distance, average delay, and number of flights.
Filter to remove noisy points and Honolulu airport, which is almost twice as far away as the next closest airport.
Power of the pipe %>% operator
# create dataframe of "delays"delays <- flights %>% group_by(dest) %>% summarize( count = n(), dist = mean(distance, na.rm = TRUE), delay = mean(arr_delay, na.rm = TRUE) ) %>% filter(count > 20, dest != "HNL")If you use RStudio, you can type the pipe with Ctrl + Shift + M if you have a PC or Cmd + Shift + M if you have a Mac.
Power of the pipe %>% operator
# create dataframe of "delays"delays <- flights %>% group_by(dest) %>% summarize( count = n(), dist = mean(distance, na.rm = TRUE), delay = mean(arr_delay, na.rm = TRUE) ) %>% filter(count > 20, dest != "HNL")If you use RStudio, you can type the pipe with Ctrl + Shift + M if you have a PC or Cmd + Shift + M if you have a Mac.
You can read it as a series of imperative statements: group, then summarize, then filter. A good way to pronounce %>% when reading code is "then".
The n() function is implemented specifically for each data source and can be used from within summarize(), mutate() and filter(). It returns the number of observations in the current group.
Transformations
Why na.rm = TRUE ? Aggregation functions obey the usual rule of missing values: if there is any missing value in the input, the output will be a missing value!
flights %>% group_by(year, month, day) %>% summarize(mean = mean(dep_delay, na.rm = TRUE))